Classes and functions related to genomics and proteomics. More...
Classes | |
| class | BamFile |
| class | InBamFile |
| class | OutBamFile |
| class | BamHeader |
| Wrapper around bam_hdr_t struct. More... | |
| class | BamPair |
| class | BamPairProxy |
| class | BamPairIterator |
| class | BamRead |
| Class holding a bam query. More... | |
| struct | BamLessPos |
| struct | BamLessEnd |
| struct | BamLessName |
| class | BamReadFilter |
| Filter bam reads. More... | |
| class | BamReadIterator |
| class to iterate through a InBamFile More... | |
| class | BamWriter |
| class | BamWriteIterator |
| Output iterator for bam file. More... | |
| class | Codon |
| struct | AminoAcidEqual |
| Functor comparing if two Codons translate to the same amino acid. More... | |
| class | DNA |
| class | GenomicPosition |
| class | GFF |
| class | GFF2 |
| class | GFF3 |
| class | Pileup |
Functions | |
| template<class Iterator , class Visitor > | |
| void | bam_pair_analyse (Iterator first, Iterator last, Visitor &visitor) |
| const unsigned char | nt16_table (char) |
| const char | nt16_str (uint8_t) |
| template<typename RandomAccessIterator > | |
| double | alignment_score (const BamRead &bam, RandomAccessIterator ref, double mismatch, double gap=0.0, double open_gap=0.0) |
| unsigned short | chr2int (const std::string &str) |
| transform a string to unsigned short chromosome number More... | |
| double | phred (double p) |
| double | phred_inv (double x) |
Detailed Description
Classes and functions related to genomics and proteomics.
Function Documentation
| double theplu::yat::omic::alignment_score | ( | const BamRead & | bam, |
| RandomAccessIterator | ref, | ||
| double | mismatch, | ||
| double | gap = 0.0, |
||
| double | open_gap = 0.0 |
||
| ) |
Calculate alignment score based on bam's cigar and sequence and the reference as in [ref, ref + bam.end()-bam.pos()).
For indels the score is calculated as open_gap + len * gap. For mismatches the score is calculated -mismatch. For matches the score is unity.
For cigar element BAM_CMATCH the sequence and reference is used to differentiate match from mismatch.
- Since
- New in yat 0.13
| void theplu::yat::omic::bam_pair_analyse | ( | Iterator | first, |
| Iterator | last, | ||
| Visitor & | visitor | ||
| ) |
bam_pair_analyse performs an operation on bam read pairs as defined by visitor. The function iterates over sorted input range of reads; if read is first read, it is cached for later use; if read is second read and mate is present in cache, visitor operates on pair, i.e., Visitor (mate, read) is called.
Type Requirements:
Iteratormust be an Input IteratorIterator'sreferencetype must be convertible to BamReadVisitormust have anoperator()(BamRead, BamRead)(or anyconstor reference combination)
- Note
- Input range
[first, last) must be sorted or behaviour is undefined.
- Since
- New in yat 0.10
| unsigned short theplu::yat::omic::chr2int | ( | const std::string & | str | ) |
transform a string to unsigned short chromosome number
If str starts with 'chr' that prefix is stripped away before translating the string to chromosome number. Function translates "X" to 23, "Y" to 24, and "M" or "MT" to 25. For other inputs utility::convert is used to transform the input to an unsigned short.
- Returns
- chromosome number
- Since
- New in yat 0.7
| const char theplu::yat::omic::nt16_str | ( | uint8_t | ) |
Convert a 4-bit encoded nucleotide to a character.
This is the same functionality that is provided in HTSLIB's global array seq_nt16_str, but this function works also when building against libbam. In libbam the array was called bam_nt16_rev_table.
This is the inverse of function nt16_table.
- Since
- New in yat 0.13
| const unsigned char theplu::yat::omic::nt16_table | ( | char | ) |
Convert nucleotide character to 4-bit encoding. The result is encoded as 1/2/4/8 for A/C/G/T or combinations of these bits.
This is the same functionality that is provided in HTSLIB's global array seq_nt16_table, but this function works also when building against libbam. In libbam the array was called bam_nt16_table.
This is the inverse of function nt16_str.
- Since
- New in yat 0.13
| double theplu::yat::omic::phred | ( | double | p | ) |
Phred scaled probability is defined as 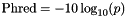
- Returns
- phred scaled probability
- Since
- New in yat 0.13
- See Also
- phred_inv
| double theplu::yat::omic::phred_inv | ( | double | x | ) |
The inverse of Phred scaling is calculated as 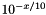
- Returns
- inverse of Phred scaling
- Since
- New in yat 0.13
- See Also
- phred
 1.8.5
1.8.5